线图
线图是反映趋势变化的一种方式,其输入数据一般也是一个矩阵。
单线图
假设有这么一个矩阵,第一列为转录起始位点及其上下游5 kb的区域,第二列为H3K27AC修饰在这些区域的丰度,想绘制一张线图展示。
profile="Pos;H3K27ac
-5000;8.7
-4000;8.4
-3000;8.3
-2000;7.2
-1000;3.6
0;3.6
1000;7.1
2000;8.2
3000;8.4
4000;8.5
5000;8.5"
读入数据 (经过前面几篇的联系,这应该都很熟了)
profile_text <- read.table(text=profile, header=T, row.names=1, quote="",sep=";")
profile_text
H3K27ac
-5000 8.7
-4000 8.4
-3000 8.3
-2000 7.2
-1000 3.6
0 3.6
1000 7.1
2000 8.2
3000 8.4
4000 8.5
5000 8.5
# 在melt时保留位置信息
# melt格式是ggplot2画图最喜欢的格式
# 好好体会下这个格式,虽然多占用了不少空间,但是确实很方便
# 这里可以用 `xvariable`,也可以是其它字符串,但需要保证后面与这里的一致
# 因为这一列是要在X轴显示,所以起名为`xvariable`。
profile_text$xvariable = rownames(profile_text)
library(ggplot2)
library(reshape2)
data_m <- melt(profile_text, id.vars=c("xvariable"))
data_m
xvariable variable value
1 -5000 H3K27ac 8.7
2 -4000 H3K27ac 8.4
3 -3000 H3K27ac 8.3
4 -2000 H3K27ac 7.2
5 -1000 H3K27ac 3.6
6 0 H3K27ac 3.6
7 1000 H3K27ac 7.1
8 2000 H3K27ac 8.2
9 3000 H3K27ac 8.4
10 4000 H3K27ac 8.5
11 5000 H3K27ac 8.5
然后开始画图,与上面画heatmap一样。
# variable和value为矩阵melt后的两列的名字,内部变量, variable代表了点线的属性,value代表对应的值。
p <- ggplot(data_m, aes(x=xvariable, y=value),color=variable) + geom_line()
p
# 图会存储在当前目录的Rplots.pdf文件中,如果用Rstudio,可以不运行dev.off()
dev.off()
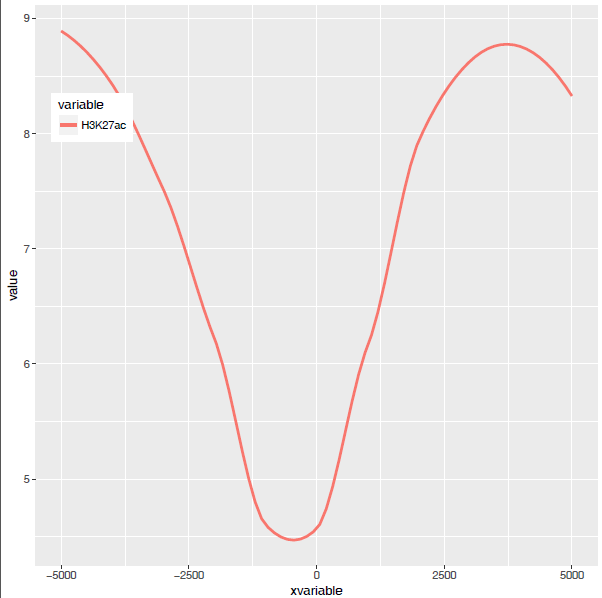
满心期待一个倒钟形曲线,结果,
什么也没有。
仔细看,出来一段提示
geom_path: Each group consists of only one observation.
Do you need to adjust the group aesthetic?
原来默认ggplot2把每个点都视作了一个分组,什么都没画出来。而data_m中的数据都来源于一个分组H3K27ac,分组的名字为variable,修改下脚本,看看效果。
p <- ggplot(data_m, aes(x=xvariable, y=value,color=variable,group=variable)) +
geom_line() + theme(legend.position=c(0.1,0.9))
p
dev.off()
图出来了,一条线,看一眼没问题;再仔细看,不对了,怎么还不是倒钟形,原来横坐标错位了。
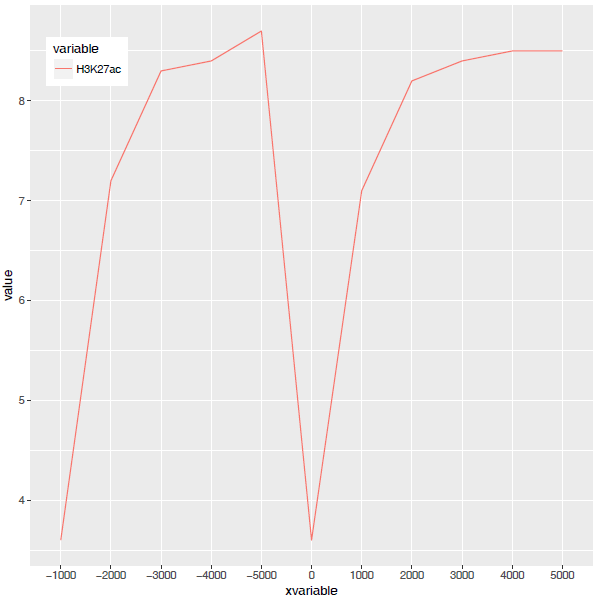
检查下数据格式
summary(data_m)
xvariable variable value
Length:11 H3K27ac:11 Min. :3.600
Class :character 1st Qu.:7.150
Mode :character Median :8.300
Mean :7.318
3rd Qu.:8.450
Max. :8.700
问题来了,xvariable虽然看上去数字,但存储的实际是字符串 (因为是作为行名字读取的),需要转换为数字。
data_m$xvariable <- as.numeric(data_m$xvariable)
#再检验下
is.numeric(data_m$xvariable)
[1] TRUE
好了,继续画图。
# 注意断行时,加号在行尾,不能放在行首
p <- ggplot(data_m, aes(x=xvariable, y=value,color=variable,group=variable)) +
geom_line() + theme(legend.position=c(0.1,0.8))
p
dev.off()
图终于出来了,调了下legend的位置,看上去有点意思了。
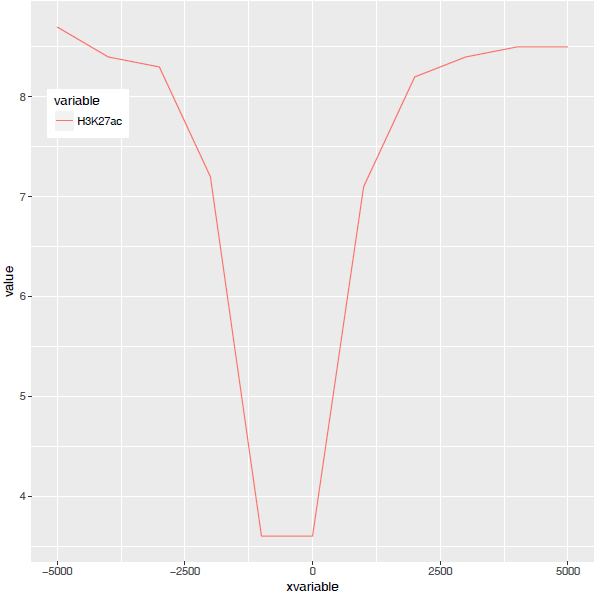
有点难看,如果平滑下,会不会好一些,stat_smooth可以对绘制的线进行局部拟合。在不影响变化趋势的情况下,可以使用 (但慎用)。
p <- ggplot(data_m, aes(x=xvariable, y=value,color=variable,group=variable)) +
geom_line() + stat_smooth(method="auto", se=FALSE) +
theme(legend.position=c(0.1,0.8))
p
dev.off()
从图中看,趋势还是一致的,线条更优美了。另外一个方式是增加区间的数量,线也会好些,而且更真实。
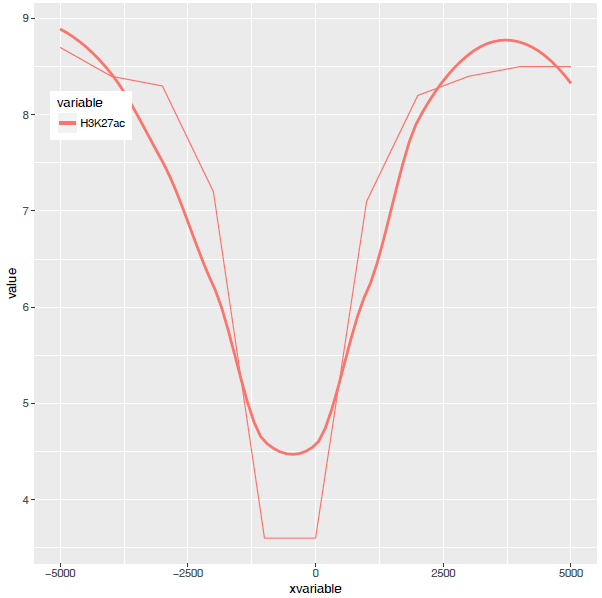
stat_smooth和geom_line各绘制了一条线,只保留一条就好。
p <- ggplot(data_m, aes(x=xvariable, y=value,color=variable,group=variable)) +
stat_smooth(method="auto", se=FALSE) + theme(legend.position=c(0.1,0.8))
p
dev.off()
好了,终于完成了单条线图的绘制。
多线图
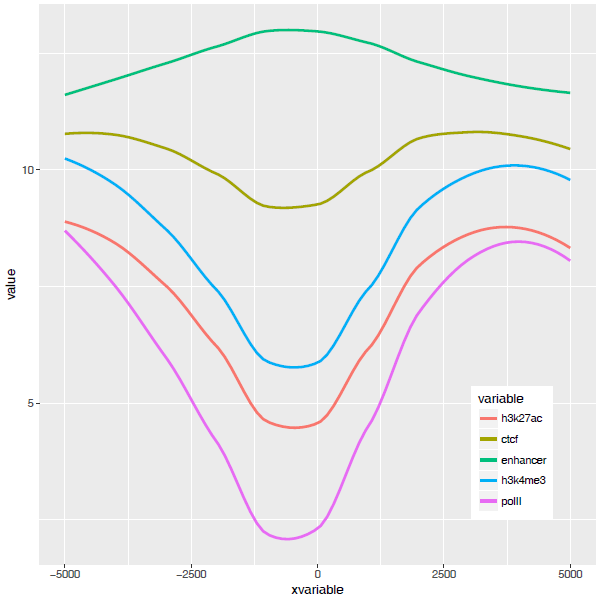
那么再来一个多线图的例子吧,只要给之前的数据矩阵多加几列就好了。
profile = "Pos;h3k27ac;ctcf;enhancer;h3k4me3;polII
-5000;8.7;10.7;11.7;10;8.3
-4000;8.4;10.8;11.8;9.8;7.8
-3000;8.3;10.5;12.2;9.4;7
-2000;7.2;10.9;12.7;8.4;4.8
-1000;3.6;8.5;12.8;4.8;1.3
0;3.6;8.5;13.4;5.2;1.5
1000;7.1;10.9;12.4;8.1;4.9
2000;8.2;10.7;12.4;9.5;7.7
3000;8.4;10.4;12;9.8;7.9
4000;8.5;10.6;11.7;9.7;8.2
5000;8.5;10.6;11.7;10;8.2"
profile_text <- read.table(text=profile, header=T, row.names=1, quote="",sep=";")
profile_text$xvariable = rownames(profile_text)
data_m <- melt(profile_text, id.vars=c("xvariable"))
data_m$xvariable <- as.numeric(data_m$xvariable)
# 这里group=variable,而不是group=1 (如果上面你用的是1的话)
# variable和value为矩阵melt后的两列的名字,内部变量, variable代表了点线的属性,value代表对应的值。
p <- ggplot(data_m, aes(x=xvariable, y=value,color=variable,group=variable)) +
stat_smooth(method="auto", se=FALSE) + theme(legend.position=c(0.85,0.2))
p
dev.off()
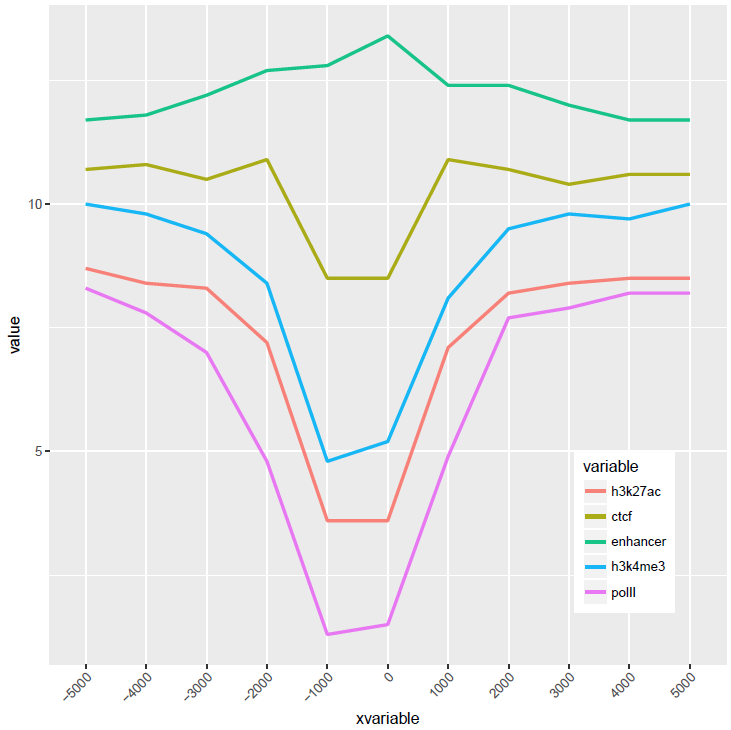
横轴文本线图
如果横轴是文本,又该怎么调整顺序呢?还记得之前热图旁的行或列的顺序调整吗?重新设置变量的factor水平就可以控制其顺序。
profile = "Pos;h3k27ac;ctcf;enhancer;h3k4me3;polII
-5000;8.7;10.7;11.7;10;8.3
-4000;8.4;10.8;11.8;9.8;7.8
-3000;8.3;10.5;12.2;9.4;7
-2000;7.2;10.9;12.7;8.4;4.8
-1000;3.6;8.5;12.8;4.8;1.3
0;3.6;8.5;13.4;5.2;1.5
1000;7.1;10.9;12.4;8.1;4.9
2000;8.2;10.7;12.4;9.5;7.7
3000;8.4;10.4;12;9.8;7.9
4000;8.5;10.6;11.7;9.7;8.2
5000;8.5;10.6;11.7;10;8.2"
profile_text <- read.table(text=profile, header=T, row.names=1, quote="",sep=";")
profile_text_rownames <- row.names(profile_text)
profile_text$xvariable = rownames(profile_text)
data_m <- melt(profile_text, id.vars=c("xvariable"))
data_m$xvariable <- factor(data_m$xvariable, levels=profile_text_rownames, ordered=T)
# geom_line设置线的粗细和透明度
p <- ggplot(data_m, aes(x=xvariable, y=value,color=variable,group=variable)) +
geom_line(size=1, alpha=0.9) + theme(legend.position=c(0.85,0.2)) +
theme(axis.text.x=element_text(angle=45,hjust=1, vjust=1))
# stat_smooth
#p <- ggplot(data_m, aes(x=xvariable, y=value,color=variable,group=variable)) +
# stat_smooth(method="auto", se=FALSE) + theme(legend.position=c(0.85,0.2)) +
# theme(axis.text.x=element_text(angle=45,hjust=1, vjust=1))
p
dev.off()
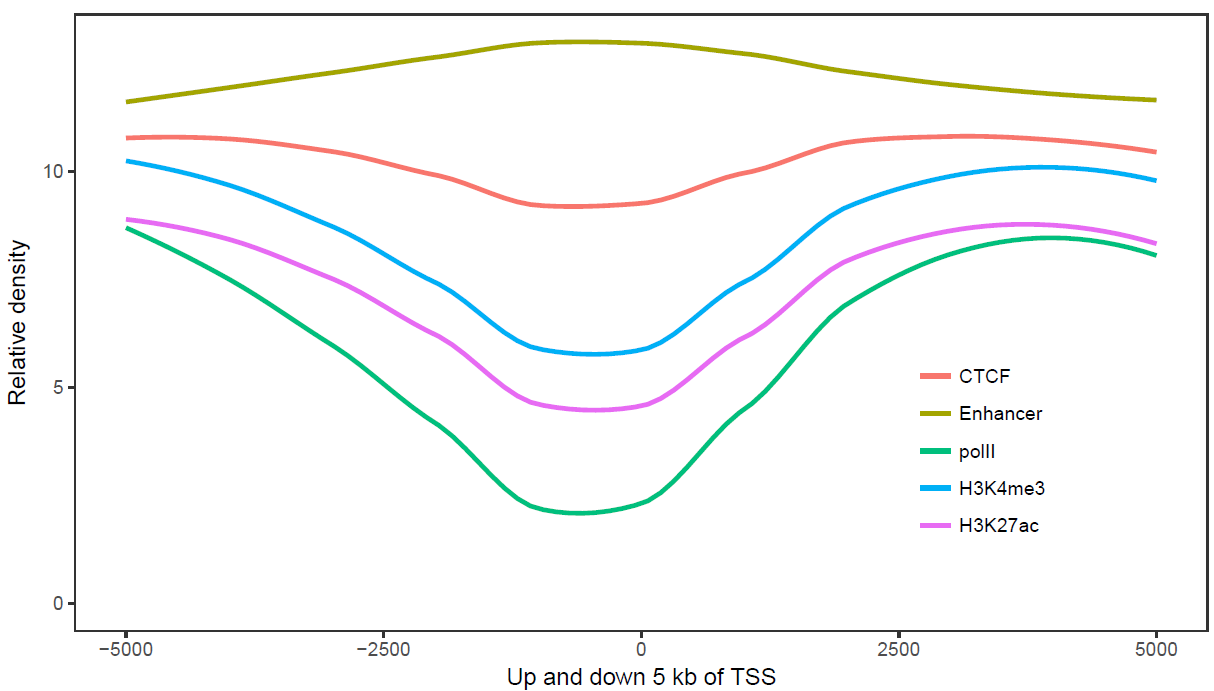
比较下位置信息做为数字(前面的线图)和位置信息横轴的差别。当为数值时,ggplot2会选择合适的几个刻度做标记,当为文本时,会全部标记。另外文本横轴,smooth效果不明显 (下面第2张图)。
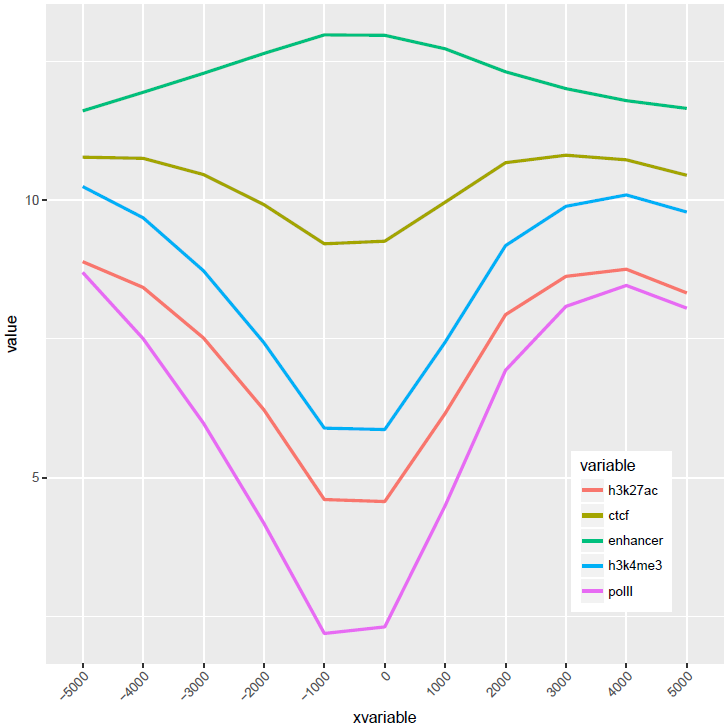
至此完成了线图的基本绘制,虽然还可以,但还有不少需要提高的地方,比如在线图上加一条或几条垂线、加个水平线、修改X轴的标记(比如0换为TSS)、设置每条线的颜色等。具体且听下回一步线图法。
线图 - 一步绘制
绘图时通常会碰到两个头疼的问题:
-
有时需要绘制很多的图,唯一的不同就是输入文件,其它都不需要修改。如果用R脚本,需要反复替换文件名,繁琐又容易出错。 (R也有命令行参数,不熟,有经验的可以尝试下)
-
每次绘图都需要不断的调整参数,时间久了不用,就忘记参数怎么设置了;或者调整次数过多,有了很多版本,最后不知道用哪个了。
为了简化绘图、维持脚本的一致,我用bash对绘图命令做了一个封装,通过配置修改命令行参数,生成相应的绘图脚本,然后再绘制。
首先把测试数据存储到文件中方便调用。数据矩阵存储在line_data.xls和line_data_melt.xls文件中 (直接拷贝到文件中也可以,这里这么操作只是为了随文章提供个测试文件,方便使用。如果你手上有自己的数据,也可以拿来用)。
profile = "Pos;H3K27ac;CTCF;Enhancer;H3K4me3;polII
-5000;8.7;10.7;11.7;10;8.3
-4000;8.4;10.8;11.8;9.8;7.8
-3000;8.3;10.5;12.2;9.4;7
-2000;7.2;10.9;12.7;8.4;4.8
-1000;3.6;8.5;12.8;4.8;1.3
0;3.6;8.5;13.4;5.2;1.5
1000;7.1;10.9;12.4;8.1;4.9
2000;8.2;10.7;12.4;9.5;7.7
3000;8.4;10.4;12;9.8;7.9
4000;8.5;10.6;11.7;9.7;8.2
5000;8.5;10.6;11.7;10;8.2"
profile_text <- read.table(text=profile, header=T, row.names=1, quote="",sep=";")
# tab键分割,每列不加引号
write.table(profile_text, file="line_data.xls", sep="\t", row.names=T, col.names=T,quote=F)
# 如果看着第一行少了ID列不爽,可以填补下
system("sed -i '1 s/^/ID\t/' line_data.xls")
profile = "Pos;variable;value;set
-5000;H3K27ac;8.71298;A
-4000;H3K27ac;8.43246;A
-3000;H3K27ac;8.25497;A
-2000;H3K27ac;7.16265;A
-1000;H3K27ac;3.55341;A
0;H3K27ac;3.5503;A
1000;H3K27ac;7.07502;A
2000;H3K27ac;8.24328;A
3000;H3K27ac;8.43869;A
4000;H3K27ac;8.48877;A
-5000;CTCF;10.6913;A
-4000;CTCF;10.7668;A
-3000;CTCF;10.5441;A
-2000;CTCF;10.8635;A
-1000;CTCF;8.45751;A
0;CTCF;8.50316;A
1000;CTCF;10.9143;A
2000;CTCF;10.7022;A
3000;CTCF;10.4101;A
4000;CTCF;10.5757;A
-5000;H3K27ac;8.71298;B
-4000;H3K27ac;8.43246;B
-3000;H3K27ac;8.25497;B
-2000;H3K27ac;7.16265;B
-1000;H3K27ac;3.55341;B
0;H3K27ac;3.5503;B
1000;H3K27ac;7.07502;B
2000;H3K27ac;8.24328;B
3000;H3K27ac;8.43869;B
4000;H3K27ac;8.48877;B
-5000;CTCF;10.6913;B
-4000;CTCF;10.7668;B
-3000;CTCF;10.5441;B
-2000;CTCF;10.8635;B
-1000;CTCF;8.45751;B
0;CTCF;8.50316;B
1000;CTCF;10.9143;B
2000;CTCF;10.7022;B
3000;CTCF;10.4101;B
4000;CTCF;10.5757;B"
profile_text <- read.table(text=profile, header=T, quote="",sep=";")
# tab键分割,每列不加引号
write.table(profile_text, file="line_data_melt.xls", sep="\t", row.names=T, col.names=T,quote=F)
# 如果看着第一行少了ID列不爽,可以填补下
system("sed -i '1 s/^/ID\t/' line_data_melt.xls")
使用正常矩阵默认参数绘制个线图
# -f: 指定输入的矩阵文件,第一列为行名字,第一行为header
列数不限,列名字不限;行数不限,行名字默认为文本
# -A FALSE: 指定行名为数字
sp_lines.sh -f line_data.xls -A FALSE
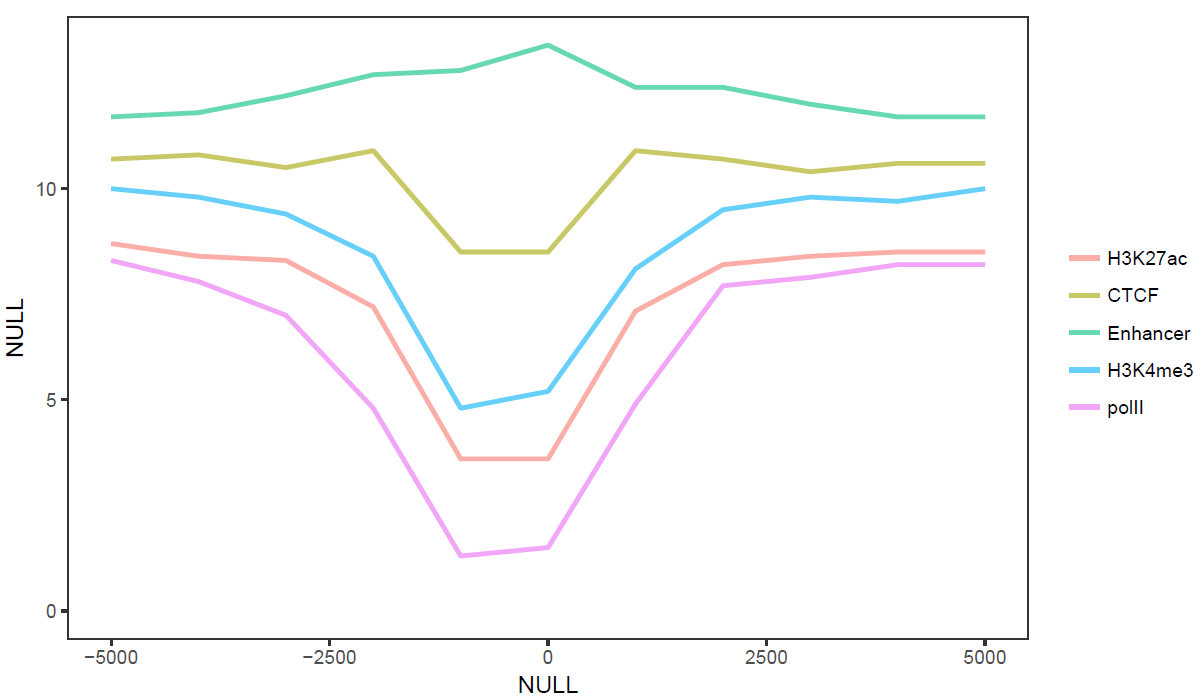
# -l: 设定图例的顺序
# -o TRUE: 局部拟合获得平滑曲线
# -A FALSE: 指定行名为数字
# -P: 设置legend位置,相对于原点的坐标
# -x, -y指定横纵轴标记
sp_lines.sh -f line_data.xls -l "'CTCF','Enhancer','polII','H3K4me3','H3K27ac'" -P 'c(0.8,0.3)' -o TRUE -A FALSE -x 'Up and down 5 kb of TSS' -y 'Relative density'
# -A FALSE: 指定行名为数字
# -V 'c(-1000, 500)': 设置垂线的位置
# -D: 设置垂线的文本标记,参数为引号引起来的vector,注意引号的嵌套
# -I: 设置横轴的标记的位置
# -b: 设置横轴标记的文字
sp_lines.sh -f line_data.xls -A FALSE -V 'c(-1000,500)' -D "c('+1 kb','-0.5 kb')" -I "c(-5000,0,5000)" -b "c('-5 kb', 'TSS', '+5 kb')"

使用melted矩阵默认参数绘制个线图 (除需要改变文件格式,指定-m TRUE, -a xvariable外其它与正常矩阵一样)
# -f: 指定输入文件
# -m TRUE: 指定输入的矩阵为melted format, 三列,第一列为Pos (给-a)
# 第二列为variable (给-H,-H默认即为variable)
# 第三列为value,名字不可修改
# -A FALSE: 指定行名为数字
# -P 'c(0.8,0.2)': 设置legend位置,相对于原点的坐标
sp_lines.sh -f line_data_melt.xls -a Pos -m TRUE -A FALSE -P 'c(0.8,0.2)'

完整的图
# -C: 自定义线的颜色
sp_lines.sh -f line_data_melt.xls -a Pos -m TRUE -A FALSE -P 'c(0.8,0.2)' -o TRUE -V 'c(-1000,500)' -D "c('+1 kb','-0.5 kb')" -I "c(-5000,0,4000)" -b "c('-5 kb', 'TSS', '+4 kb')" -x 'Up 5 kb and down 4 kb of TSS' -y 'Relative density' -C "'pink', 'blue'"
参数中最需要注意的是引号的使用:
- 外层引号与内层引号不能相同
- 凡参数值中包括了
空格,括号,逗号等都用引号括起来作为一个整体。
完整参数列表如下:
ct@ehbio:~ $sp_lines.sh
***CREATED BY Chen Tong (chentong_biology@163.com)***
Usage:
/MPATHB/self/s-plot/sp_lines.sh options
Function:
This script is used to draw a line or multiple lines using ggplot2.
You can specify whether or not smooth your line or lines.
Two types of input files are supported, normal matrix or melted matrix format. Column separator for both types of input files is **tab**.
Here is an example of normal matrix format. The first column will be treated as X-axis variables and other columns represents each type of lines. The number of columns is unlimited and names of columns is unlimited.
**Set** column is not needed. If given, <facet_plot> (multiple plots in one page) could be displayed.
------------------------------------------------------------
Pos H3K27ac CTCF Enhancer H3K4me3 polII
-5000 8.71298 10.69130 11.7359 10.02510 8.26866
-4000 8.43246 10.76680 11.8442 9.76927 7.78358
-3000 8.25497 10.54410 12.2470 9.40346 6.96859
-2000 7.16265 10.86350 12.6889 8.35070 4.84365
-1000 3.55341 8.45751 12.8372 4.84680 1.26110
0 3.55030 8.50316 13.4152 5.17401 1.50022
1000 7.07502 10.91430 12.3588 8.13909 4.88096
2000 8.24328 10.70220 12.3888 9.47255 7.67968
3000 8.43869 10.41010 11.9760 9.80665 7.94148
4000 8.48877 10.57570 11.6562 9.71986 8.17849
------------------------------------------------------
------------With SET------------------------------------------
Pos H3K27ac CTCF Enhancer H3K4me3 polII Set
-5000 8.71298 10.69130 11.7359 10.02510 8.26866 1
-4000 8.43246 10.76680 11.8442 9.76927 7.78358 1
-3000 8.25497 10.54410 12.2470 9.40346 6.96859 1
-2000 7.16265 10.86350 12.6889 8.35070 4.84365 1
-1000 3.55341 8.45751 12.8372 4.84680 1.26110 1
0 3.55030 8.50316 13.4152 5.17401 1.50022 1
1000 7.07502 10.91430 12.3588 8.13909 4.88096 1
2000 8.24328 10.70220 12.3888 9.47255 7.67968 1
3000 8.43869 10.41010 11.9760 9.80665 7.94148 1
4000 8.48877 10.57570 11.6562 9.71986 8.17849 1
-5000 8.71298 10.69130 11.7359 10.02510 8.26866 2
-4000 8.43246 10.76680 11.8442 9.76927 7.78358 2
-3000 8.25497 10.54410 12.2470 9.40346 6.96859 2
-2000 7.16265 10.86350 12.6889 8.35070 4.84365 2
-1000 3.55341 8.45751 12.8372 4.84680 1.26110 2
0 3.55030 8.50316 13.4152 5.17401 1.50022 2
1000 7.07502 10.91430 12.3588 8.13909 4.88096 2
2000 8.24328 10.70220 12.3888 9.47255 7.67968 2
3000 8.43869 10.41010 11.9760 9.80665 7.94148 2
4000 8.48877 10.57570 11.6562 9.71986 8.17849 2
-------------------------------------------------------------
For matrix format, example command lines include:
* Attribute of X-axis value (first column of matrix) is <number>
s-plot lines -f matrix.file -A FALSE
* Attribute of X-axis value (first column of matrix) is <text>
s-plot lines -f matrix.file
* Attribute of X-axis value (first column of matrix) is numbers, change legned order (default alphabet order)
s-plot lines -f matrix.file -l "'polII', 'CTCF', 'Enhancer', 'H3K27ac', 'H3K4me3'"
* Attribute of X-axis value (first column of matrix) is numbers, change legned order (default alphabet order), smooth lines to look better (Pay attention to whether this will change the data trend)
s-plot lines -f matrix.file -l "'polII', 'CTCF', 'Enhancer', 'H3K27ac', 'H3K4me3'" -o TRUE
* Attribute of X-axis value (first column of matrix) is numbers, with <Set> (Set is column name) column
s-plot lines -f matrix.file -F "+facet_grid(Set ~ ., scale='free_y')"
FILEFORMAT when -m is true
#The name "value" shoud **not** be altered.
#variable can be altered using -H
#Actually this format is the melted result of last format.
--------------------------------------------------------------
Pos variable value
-5000 H3K27ac 8.71298
-4000 H3K27ac 8.43246
-3000 H3K27ac 8.25497
-2000 H3K27ac 7.16265
-1000 H3K27ac 3.55341
0 H3K27ac 3.55030
1000 H3K27ac 7.07502
2000 H3K27ac 8.24328
3000 H3K27ac 8.43869
4000 H3K27ac 8.48877
-5000 CTCF 10.69130
-4000 CTCF 10.76680
-3000 CTCF 10.54410
-2000 CTCF 10.86350
-1000 CTCF 8.45751
0 CTCF 8.50316
1000 CTCF 10.91430
2000 CTCF 10.70220
3000 CTCF 10.41010
4000 CTCF 10.57570
-------------------------------------------------------------
* Attribute of X-axis value (melt format) is <number>
s-plot lines -f matrix.file -m TRUE -a Pos -A FALSE
* Attribute of X-axis value (first column of matrix) is <text>
s-plot lines -f matrix.file -m TRUE -a Pos
* If the name of the second column is <type> not <variable>, one should specify with <-H>.
s-plot lines -f matrix.file -A FALSE -m TRUE -a Pos -H type
* Attribute of X-axis value (first column of matrix) is numbers, change legned order (default alphabet order)
s-plot lines -f matrix.file -m TRUE -a Pos -l "'polII', 'CTCF', 'Enhancer', 'H3K27ac', 'H3K4me3'"
* Attribute of X-axis value (first column of matrix) is numbers, change legned order (default alphabet order), smooth lines to look better (Pay attention to whether this will change the data trend)
s-plot lines -f matrix.file -m TRUE -a Pos -l "'polII', 'CTCF', 'Enhancer', 'H3K27ac', 'H3K4me3'" -o TRUE
* Attribute of X-axis value (first column of matrix) is numbers, with <Set> (Set is column name) column
s-plot lines -f matrix.file -F "+facet_grid(Set ~ ., scale='free_y')"
OPTIONS:
-f Data file (with header line, the first column would be be treated as rownames for
normal matrix. No rownames for melted format. Columns are tab seperated)
[NECESSARY]
-m When true, it will skip melt preprocesses. But the format must be
the same as listed before.
[Default FALSE, accept TRUE]
-a Name for x-axis variable
[Only needed when <-m> is <TRUE>.
For the melted data, 'Pos' should be given here.
For normal matrix, default the first column will be used,
program will assign an value 'xvariable' to represent it.
]]
-A Are x-axis variables numbers.
[Default <TRUE>, meaning X-axis label is <text>.
<FALSE> means X-axis label is <numerical>.]
-H Name for legend variable.
[Default variable, this should only be set when -m is TRUE]
-J Name for color variable.
[Default same as -H, this should only be set when -m is TRUE]
-l Set orders of legend variable.
[Default column order for normal matrix, accept a string like
"'CTCF','H3K27ac','Enhancer'" to set your own order.
Pay attention to the usage of two types of quotes.
***When -m is TRUE, default order would be alphabet order.*********
]
-P Legend position[Default right. Accept
top, bottom, left, none, or 'c(0.08,0.8)'.]
-L Levels for x-axis variable, suitable when x-axis is not treated as numerical.
[Default the order of first column for normal matrix.
Accept a string like "'g','a','j','x','s','c','o','u'" to set your own oder.
This will only be considered when -A is TRUE.
***When -m is used, this default order would be alphabet order.*********
]
-o Smooth lines or not.
[Default FALSE means no smooth. Accept TRUE to smooth lines.]
-O The smooth method you want to use.
[smoothing method (function) to use, eg. lm, glm, gam, loess,rlm.
For datasets with n < 1000 default is 'loess'.
For datasets with 1000 or more observations defaults to 'gam'.
]
-V Add vertical lines.[Default FALSE, accept a series of
numbers in following format "c(1,2,3,4,5)" or other
R code that can generate a vector.]
-D Add labels to vlines.
[Default same as -V.
Accept a series of numbers in following format "c(1,2,3,4,5)" or other R code
that can generate a vector as labels.
Or one can give '1' to disallow labels]
-j Add horizontal lines.[Default FALSE, accept a series of
numbers in following format "c(1,2,3,4,5)" or other
R code that can generate a vector]
-d Add labels to hline.
[Default same as -j
Accept a series of numbers in following format "c(1,2,3,4,5)" or other R code
that can generate a vector as labels.
Or one can give '1' to disallow labels]
-I Manually set the position of xtics.
[Default FALSE, accept a series of
numbers in following format "c(1,2,3,4,5)" or other R code
that can generate a vector to set the position of xtics]
-b Manually set the value of xtics when -I is specified.
[Default the content of -I when -I is specified,
accept a series of numbers in following format "c(1,2,3,4,5)" or other R code
that can generate a vector to set the position of xtics]
-X Display xtics. [Default TRUE]
-Y Display ytics. [Default TRUE]
-R Rotation angle for x-axis labels (anti clockwise)
[Default 0]
-B line size. [Default 1. Accept a number.]
-t Title of picture[Default empty title]
-x xlab of picture[Default empty xlab]
-y ylab of picture[Default empty ylab]
-c Manually set colors for each line.[Default FALSE, meaning using ggplot2 default.]
-C Color for each line.
When -c is TRUE, one has two options:
1. Supplying a function to generate colors,
like "rainbow(11)" or "rainbow(11, alpha=0.6)",
rainbow is an R color palletes,
11 is the number of colors you want to get,
0.6 is the alpha value.
The R palletes include <heat.colors>, <terrain.colors>,
<topo.colors>, <cm.colors>.
2. Supplying a list of colors in given format,
the number of colors should be equal to the number of
bars like "'red','pink','blue','cyan','green','yellow'" or
"rgb(255/255,0/255,0/255),rgb(255/255,0/255,255/255),
rgb(0/255,0/255,255/255),rgb(0/255,255/255,255/255),
rgb(0/255,255/255,0/255),rgb(255/255,255/255,0/255)"
One can use R fucntion <colors()> to list all available colors.
-s Scale y axis
[Default null. Accept TRUE. This function is depleted.
But if the supplied number after -S is not 0, this parameter will be set to TRUE]
-F The formula for facets.[Default no facets,
"+facet_grid(level ~ .)" means divide by levels of 'level' vertically.
"+facet_grid(. ~ level)" means divide by levels of 'level' horizontally.
"+facet_grid(lev1 ~ lev2)" means divide by lev1 vertically and lev2 horizontally.
"+facet_wrap(~level, ncol=2)" means wrap horizontally with 2 columns.
#Pay attention to the single quote for parameters in function for scale.
Example: "+facet_wrap(~Size,ncol=6,scale='free')"
Example: "+facet_grid(Size ~ .,scale='free_y')"
]
-G If facet is given, you may want to specifize the order of
variable in your facet, default alphabetical order.
[Accept sth like (one level one sentence, separate by';')
'data$size <- factor(data$size, levels=c("l1", "l2",...,"l10"), ordered=T)' ]
-v If scale is TRUE, give the following 'scale_y_log10()'[default], 'coord_trans(y="log10")',
or other legal command for ggplot2 or simply 'log2'.]
-S A number to add if scale is used.
[Default 0. If a non-zero number is given, -s would be set to TRUE.]
-p Other legal R codes for gggplot2 could be given here.
[Begin with '+' ]
-w The width of output picture (cm).[Default 20]
-u The height of output picture (cm).[Default 12]
-E The type of output figures.[Default pdf, accept
eps/ps, tex (pictex), png, jpeg, tiff, bmp, svg and wmf)]
-r The resolution of output picture.[Default 300 ppi]
-z Is there a header. Must be TRUE. [Default TRUE]
-e Execute or not[Default TRUE]
-i Install depended packages[Default FALSE]
为了推广,也为了激起大家的热情,如果想要sp_lines.sh脚本的,还需要劳烦大家动动手,转发此文章到朋友圈,并留言索取。
也希望大家能一起开发,完善功能。
Reference
- http://blog.genesino.com//2017/06/R-Rstudio
