How to tell which library type to use (fr-firststrand or fr-secondstrand)
| Library Type | Examples | Description |
| fr-unstranded | Standard Illumina | Reads from the left-most end of the fragment (in transcript coordinates) map to the transcript strand, and the right-most end maps to the opposite strand. |
| fr-firststrand | dUTP, NSR, NNSR | Same as above except we enforce the rule that the right-most end of the fragment (in transcript coordinates) is the first sequenced (or only sequenced for single-end reads). Equivalently, it is assumed that only the strand generated during first strand synthesis is sequenced. |
| fr-secondstrand | Ligation, Standard SOLiD | Same as above except we enforce the rule that the left-most end of the fragment (in transcript coordinates) is the first sequenced (or only sequenced for single-end reads). Equivalently, it is assumed that only the strand generated during second strand synthesis is sequenced. |
In case you don’t know the library-type, you can still figure it out by yourself. Tophat FAQ page provided a solution for that (http://tophat.cbcb.umd.edu/faq.html#library_type). But more simply (comparing to running 1M reads first), you can choose few reads and BLAT to genome and infer the library-type from the mapping result.
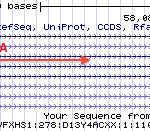
Generally, reads from the left-most end of RNA fragment (always from 5´ to 3´) are always mapped to transcript-strand, and (for pair-end sequencing) reads from the right-most end are always mapped to the opposite strand. See the arrows direction in the below schema. This is because the sequencer always read from 5´ to 3´.
|
But regarding to which strand the RNA fragment is synthesized from, this involves different strand-specific protocols. Thanks to the illustration figure (see below) from Zhao Zhang, we could see that for example dUTP method is to only sequence the strand from the first strand synthesis (the original RNA strand is degradated due to the dUTP incorporated), so the /2 read is from the original RNA strand.
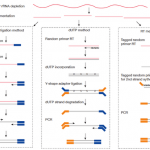
| Strand-specific library protocols (Credit: Zhao Zhang) |
Taking a real example, first getting some reads (in fasta format) from the paired-end sequencing fastq file using command like:
$ zcat ~/nearline/rnaseq/BU/Jul2012/Sample_3576_H_01.R1.fastq.gz | sed ‘s/@//g;s/ /_/g’ | awk ‘{if(NR%4==1)print “>”$0;if(NR%4==2) print $0;}’ | head
$ zcat ~/nearline/rnaseq/BU/Jul2012/Sample_3576_H_01.R2.fastq.gz | sed ‘s/@//g;s/ /_/g’ | awk ‘{if(NR%4==1)print “>”$0;if(NR%4==2) print $0;}’ | head
Blatting them in UCSC Genome Browser
Continuing to look at other reads in the file, we can find examples like these: