UCSC usages
Here lists how to use UCSC tablebrowser to get various gene annotation information.
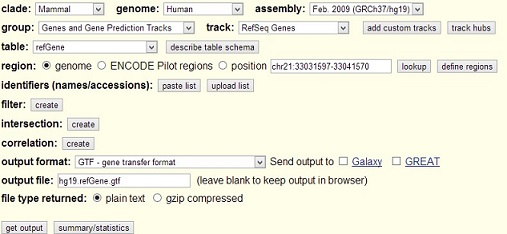
Get gene annotation file in GTF or bed format
Open UCSC table browser and fill the following information and hit get output.

Get chromosome size file or genome size file
mysql --user=genome --host=genome-mysql.cse.ucsc.edu -A -e \
"select chrom, size from hg19.chromInfo" > hg19.chromsome.size
#or
fetchChromSizes mm9 > mm9.chrom.sizesGet the type of each transcripts
- For Refseq genes
- Change
tablefromrefGenetorefSeqStatusin above picture. - Click
describe table schemato see the description of each table. Here lists the information for reference.
- Change
–Status
'Unknown', 'Reviewed', 'Validated', 'Provisional', 'Predicted', 'Inferred'
–Molecular type
'DNA', 'RNA', 'ds-RNA', 'ds-mRNA', 'ds-rRNA', 'mRNA', 'ms-DNA', 'ms-RNA', 'rRNA', 'scRNA',
'snRNA', 'snoRNA', 'ss-DNA', 'ss-RNA', 'ss-snoRNA', 'tRNA', 'cRNA', 'ss-cRNA', 'ds-cRNA', 'ms-rRNA'
- For Ensemble genes
- From track
ensemblGenesand tableensemblSource. - Click
describe table schemato see the description of each table. Here lists the information for reference.
- From track
name source
ENSMUST00000160944 transcribed_unprocessed_pseudogene
ENSMUST00000082908 snRNA
ENSMUST00000162897 processed_transcript
ENSMUST00000159265 processed_transcript
ENSMUST00000070533 protein_coding
ENSMUST00000161581 antisense
ENSMUST00000157765 snRNA
- Please also refer to http://asia.ensembl.org/info/docs/genebuild/ncrna.html and http://redmine.soe.ucsc.edu/forum/index.php?t=msg&goto=12047&S=03eba72760c0c4d83c9a2327810936cb.
Get pseudogene annotation in GTF format
Open UCSC table browser and chose alltables for group and pseudoYale60 for table.
Get rRNA annotation in GTF format
- Select “All Tables” from the group drop-down list
- Select the “rmsk” table from the table drop-down list
- Choose “GTF” as the output format
- Type a filename in “output file” so your browser downloads the result
- Click “create” next to filter
- Next to “repClass,” type rRNA
- Next to free-form query, select “OR” and type repClass = “tRNA”
- Click submit on that page, then get output on the main page
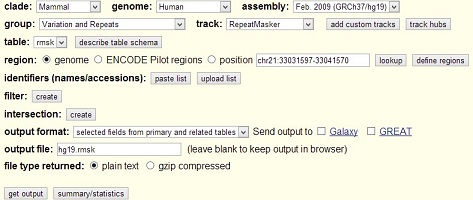
Get repeat elements and their class and family information
- UCSC table browser — mammal – mouse — mm9 - variation and repeats – repeatmarsker — rmsk – selected fields from primary and related tables
Get and use Phastcon conservation value
- Get PhastCons scores from UCSC:
There are various phastCons score files, depending on the genomes in the pileup. E.g. http://hgdownload.cse.ucsc.edu/goldenPath/mm9/phastCons30way/. You can get them by ftp:
wget ftp://hgdownload.cse.ucsc.edu/goldenPath/mm9/phastCons30way/vertebrate/chr*2) Convert wig to bigWig:
Each chromosome file of scores (wig files) can be converted into bigWig files. 64-bit binaries come from: http://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/
The fetchChromSizes script is required to get the chromosome length for the wigToBigWig script
fetchChromSizes mm9 > mm9.chrom.sizesRun wigToBigWig on each chromosome file, e.g:
wigToBigWig -clip chr1.data mm9.chrom.sizes chr1.bw3) Calculate mean phastCons score for the coordinates of interest:
Another script called bigWigSummary can be obtained from UCSC, using the link above. It takes in a set of genome coordinate and will spit out the max or mean of values contained in the bigWig file, in our case phastCons scores.
